U1160 - Team 1 “Homeostatic and Pathological Chemokine-Regulated Interplays between Lymphocytes and their Microenvironment”
 Team leader : Karl Balabanian, DR2 Inserm, Team Leader
Team leader : Karl Balabanian, DR2 Inserm, Team Leader
 Deputy team leader : Marion Espéli, CRCN Inserm, Deputy Team Leader
Deputy team leader : Marion Espéli, CRCN Inserm, Deputy Team Leader
Composition
BALABANIAN Karl (DR2 INSERM, HDR)
ESPÉLI Marion (CRCN INSERM, HDR)
ANGINOT Adrienne (CRCN INSERM, HDR in progress)
DULPHY Nicolas (MCU-PH Univ. Paris, APHP, HDR)
REA Delphine (PH APHP)
LERECLUS Emilie (IE INSERM)
BONAUD Amélie (Post-Doctorante)
LEMOS Julia (Post-Doctorante)
BISIO Valeria (Post-Doctorante)
RONDEAU Vincent (PhD student)
BOY Maxime (PhD student)
ZHAO Lin-Pierre (MD-PhD student)
ABOU NADER Zeina (PhD student)
KHAMYATH Mélanie (PhD student)
MOULIN Clémentine (Master 2 student)
SCHELL Bérénice (Master 2 student)
Main topic:
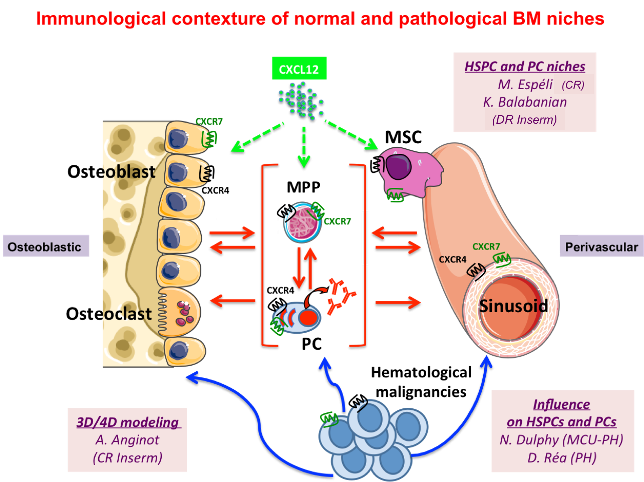
Our activities are centered on the interplays established between innate (NK, ILC) or adaptive (multipotent progenitors/MPP, plasma cells/PC) lymphoid cells and their microenvironment with a special focus on chemokine signaling in the bone marrow (BM). This is addressed at steady-state and under specific pathological conditions that arise in the BM: the WHIM Syndrome (WS), a rare immune-hematological disorder, Waldenström’s Macroglobulinemia (WM), a lymphoplasmacytic lymphoma with IgM monoclonal protein, and Myelodysplastic Syndromes (MDS) whom the evolution is dominated by the risk of progression into Acute Myeloid Leukemia (AML).
Scientific strategy:
The project is divided in three interconnected axes:
1) Cartography of BM localization and fate of MPP/PC and identification of the molecular interplays between MPP, PC and their BM niches in health and disease. Led by KB (MPP) and ME (PC).
2) Direct and stroma-mediated influence of MDS blasts on innate lymphopoiesis. Led by ND.
3) Dynamics of bone stroma and modelling of normal and pathologic HSPC and PC niches. Led by AA.
We are using a combination of classic immunological (e.g. Flow cytometry, confocal microscopy, in vitro functional assays) and cutting-edge (e.g Single-cell transcriptomics, artificial intelligence-driven image analysis, complex co-culture systems) technologies to address how lymphoid cells and their environment communicate in a bi-directional manner. We work on human samples and original genetically modified mouse models to dissect how this dialogue might be impaired in the course of immunodeficiencies and malignancies.
Our international, dynamic and enthusiastic team is always looking for new members eager to join the adventure.
Ten main outputs during the 2015-2020 period
1. BONAUD, A., Clare, S., BISIO, V., Sowerby, JM., Yao, S., Ostergaard, H., BALABANIAN, K., Smith, KGC., and ESPELI, M. (2020). Leupaxin Expression Is Dispensable for B Cell Immune Responses. Front Immunol 11, 466.
2. Boutin, L., Arnautou, P., Trignol, A., Ségot, A., Farge, T., Desterke, C., Soave, S., Clay, D., Raffoux, E., Sarry, JE., Malfuson, JV., Lataillade, JJ., Le Bousse-Kerdilès, MC., and ANGINOT, A. (2020). Mesenchymal stromal cells confer chemoresistance to myeloid leukemia blasts through Side Population functionality and ABC transporter activation. Haematologica 105, 987-9998.
3. ESPELI, M., Bashford-Rogers, R., Sowerby, JM., Alouche, N., Wong, L., Denton, AE., Linterman, MA., and Smith, KGC. (2019). FcγRIIb Differentially Regulates Pre-Immune and Germinal Center B Cell Tolerance in Mouse and Human. Nat Commun 10,1970.
4. Freitas, C., RONDEAU, V., and BALABANIAN, K. (2018). New method to obtain lymphoid progenitors. Patent WO/2018/177971 (PCT/EP2018/057580). PATENT
5. REA, D., Henry, G., Khaznadar, Z., Etienne, G., Guilhot, F., Nicolini, F., Guilhot, J., Rousselot, P., Huguet, F., Legros, L., Gardembas, M., Dubruille, V., Guerci-Bresler, A., Charbonnier, A., Maloisel, F., Ianotto, JC., Villemagne, B., Mahon, FX., Moins-Teisserenc, H., DULPHY, N., and Toubert, A. (2017). Natural killer cell counts are associated with molecular relapse-free survival after imatinib discontinuation in chronic myeloid leukemia: the IMMUNOSTIM study. Haematologica 102, 1368-1377.
6. Freitas, C., Wittner, M., Nguyen, J., RONDEAU, V., Biajoux, V., Aknin, M.L., Gaudin, F., Beaussant-Cohen, S., Bertrand, Y., Bellanne-Chantelot, C., Donadieu, J., Bachelerie, F., ESPELI, M., Dalloul, A., Louache, F.*, and BALABANIAN, K.* (2017). Lymphoid differentiation of hematopoietic stem cells requires efficient Cxcr4 desensitization. J Exp Med 214, 2023-2040.*Co-senior authors
7. REA, D., Nicolini, FE., Tulliez, M., Guilhot, F., Guilhot, J., Guerci-Bresler, A., Gardembas, M., Coiteux, V., Guillerm, G., Legros, L., Etienne, G., Pignon, JM., Villemagne, B., Escoffre-Barbe, M., Ianotto, JC., Charbonnier, A., Johnson-Ansah, H., Noel, MP., Rousselot, P., and Mahon, FX.; France Intergroupe des Leucémies Myéloïdes Chroniques. (2017). Discontinuation of dasatinib or nilotinib in chronic myeloid leukemia: interim analysis of the STOP 2G-TKI study. Blood 129, 846-854.
8. Biajoux, V., Natt, J., Freitas, C., Alouche, N., Sacquin, A., Hemon, P., Gaudin, F., Fazilleau, N., ESPELI, M.*, and BALABANIAN, K.* (2016). Efficient Plasma Cell Differentiation and Trafficking Require Cxcr4 Desensitization. Cell Rep 17, 193-205.*Co-senior authors
9. Desnoyer, A., Dupin, N., Assoumou, L., Carlotti, A., Gaudin, F., Deback, C., Peytavin, G., Marcelin, A.G., Boue, F., BALABANIAN, K.*, and Pourcher, V.* ANRS 154 LENAKAP trial group (2016). Expression pattern of the CXCL12/CXCR4-CXCR7 trio in Kaposi sarcoma skin lesions. Br J Dermatol 175, 1251-1262.*Co-senior authors
10. Bignon, A., Regent, A., Klipfel, L., Desnoyer, A., de la Grange, P., Martinez, V., Lortholary, O., Dalloul, A., Mouthon, L., and BALABANIAN, K. (2015). DUSP4-mediated accelerated T-cell senescence in idiopathic CD4 lymphopenia. Blood 125, 2507-2518.
Major grants during the 2015-2020 period
JCJC ANR 2019 PC-SEC
Ligue contre le Cancer project grant 2019
IDEX-Université de Paris “Dynamique de Recherche” 2019
Association St-Louis pour la recherche sur les Leucémies 2018
ARC project grant 2018
Fondation Arthritis 2018
POC Inserm Transfert 2017
PRC ANR 2017 Osteovalymph
PRT-K INCa/DGOS 2017 BM-Immune-MDS
Association Force Hémato 2017
Association Laurette Fugain 2016
ANR@RAction 2015 AUTO-PLASMO
Labex LERMIT “Target” and Junior Team Leader 2015
Main networks
GDR 3697 MicroNiT
IHU THEMA
Institut Carnot OPALE
FHU Prothee
FHU TRUE
COST EuNet Innochron network
Alumni
V. Biajoux (2009-13)
A. Bignon (2010-14)
C. Freitas (2012-16)
A. Desnoyer (2012-15)
J. Nguyen (2014-2018)
N. Alouche (2014-2018)
J. Natt (2014-2017)
L. Klipfel (2012-15)